Getting started
Glossary
Sample ID The identifier of each entire where the mutation identified in the reference.
Response Descript the qualitative measurement of entries. P (positive, response); N (negative,no response).
Tumor Type The tumor type of patient or cell line, such as NSC Lung cancer (non-small cell lung cancer) and SC Lung cancer (small cell lung cancer).
Gene Symbol Genetic symbols refer to the set of symbols used for the representation of various genes and include the protocol for the nomenclature of genes. In our database, it represents the gene where neoantigens came from. Each gene symbol was unified from NCBI.
FPKM Fragments per kilobase of transcript per million. This data revealed that the transcriptome expression levels in mutant genes in tumor sample.
DNA mutation Single nuclear acid substitution on gene level.
Transcript ID Corresponding transcript id of gene.
mut_AA Position The single amino acid mutation position of protein.
Antigen Type The type of antigen which used in trials. TMG (tandem minigene).
HLA-A The HLA (human leukocyte antigen) class Ⅰ loci A associated with the epitope, as described in the article.
HLA-B The HLA (human leukocyte antigen) class Ⅰ loci B associated with the epitope, as described in the article.
HLA-C The HLA (human leukocyte antigen) class Ⅰ loci C associated with the epitope, as described in the article.
T-cell Source The source where the T cells came from in immunoassay. CTL (cytotoxic T lymphocyte); PBMC (Peripheral blood mononuclear cell); TIL (tumor infiltrating lymphocyte).
APC Type Antigen presenting cell type in immunoassay. DC (dendritic cell); MoDCs (monocyte-derived dendritic cell).
Assay This list includes immunological methods, such as ELISPOT, tetramer. ELISPOT (enzyme-linked immunospot).
Checkpoint Blockade Checkpoint blockade treatment in clinical trial. (Yes/No)
ACT Adoptive cell transfer used in clinical trial. (Yes/No)
Vaccination Vaccine used in clinical trial. (Yes/No)
Curative Effect Description of clinical curative effect in the reference.
How to search the Validated Neopeptide Dataset (VND)?
The search interface is user-friendly and convenient. One or more filters can be specified, including response, specific peptide, protein sequence, tumor type, HLA type, gene symbol, and published year.
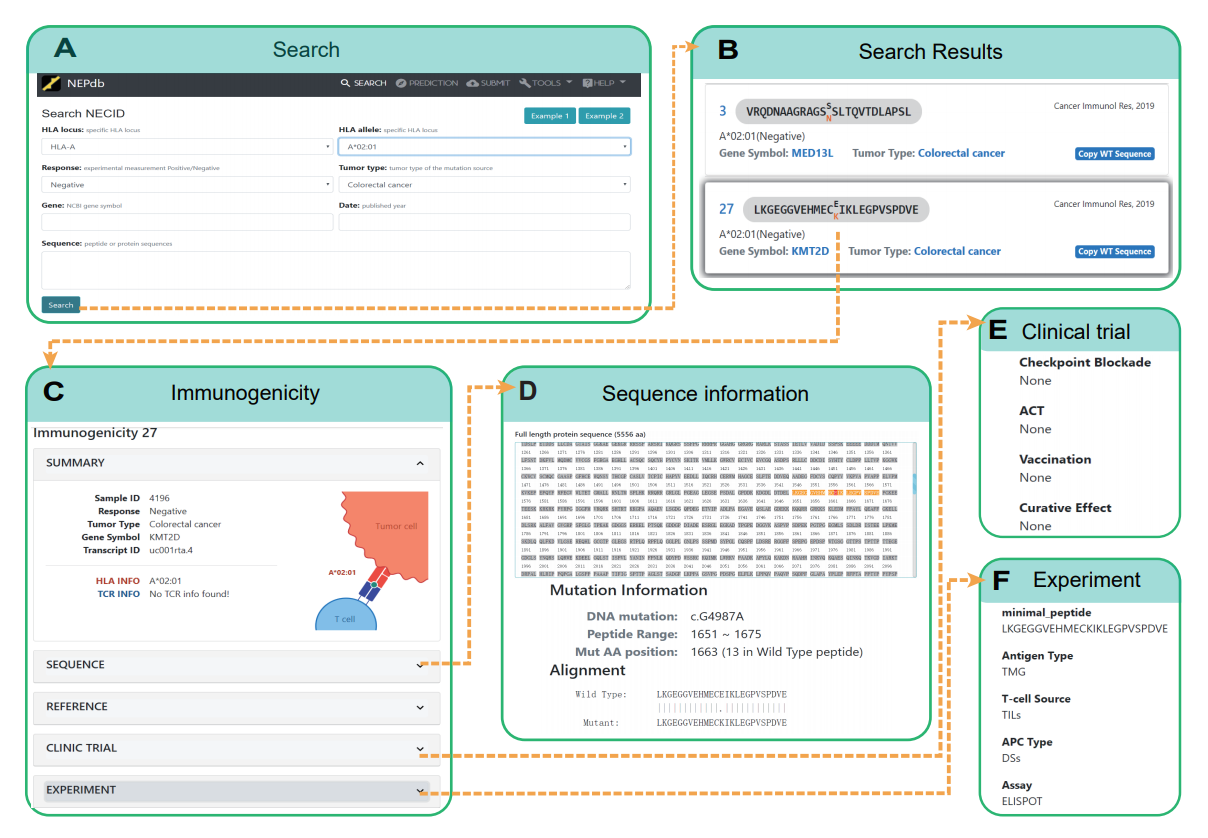
As shown above, it is a comprehensive view of NEPdb search and query results. Panels (A) options include 'HLA locus', 'Response', 'Tumor type', 'Sequence' and 'gene', click Search to jump to panel (B) show wild peptide, mutation peptide, mutation AA, gene symbol, HLA type, literature etc. Panel (C) Details of a selected peptide from the results table. Panels (D), (E) and (F) are result of full protein sequence, clinical trial and experimental information.
How to explore the Predicted Neopeptide Dataset (PND)?
A neopeptide pool was computationally created for each cancer gene derived non-synonymous mutations associated peptides potentially binding to HLA-I. For each mutation, we performed a comprehensive assessment of peptides 8–11 amino acids in length at every position surrounding a somatic mutation. We predicted the binding potentials with 95 HLA class I alleles using NetMHCpan 4.0 and HLAthena , respectively.
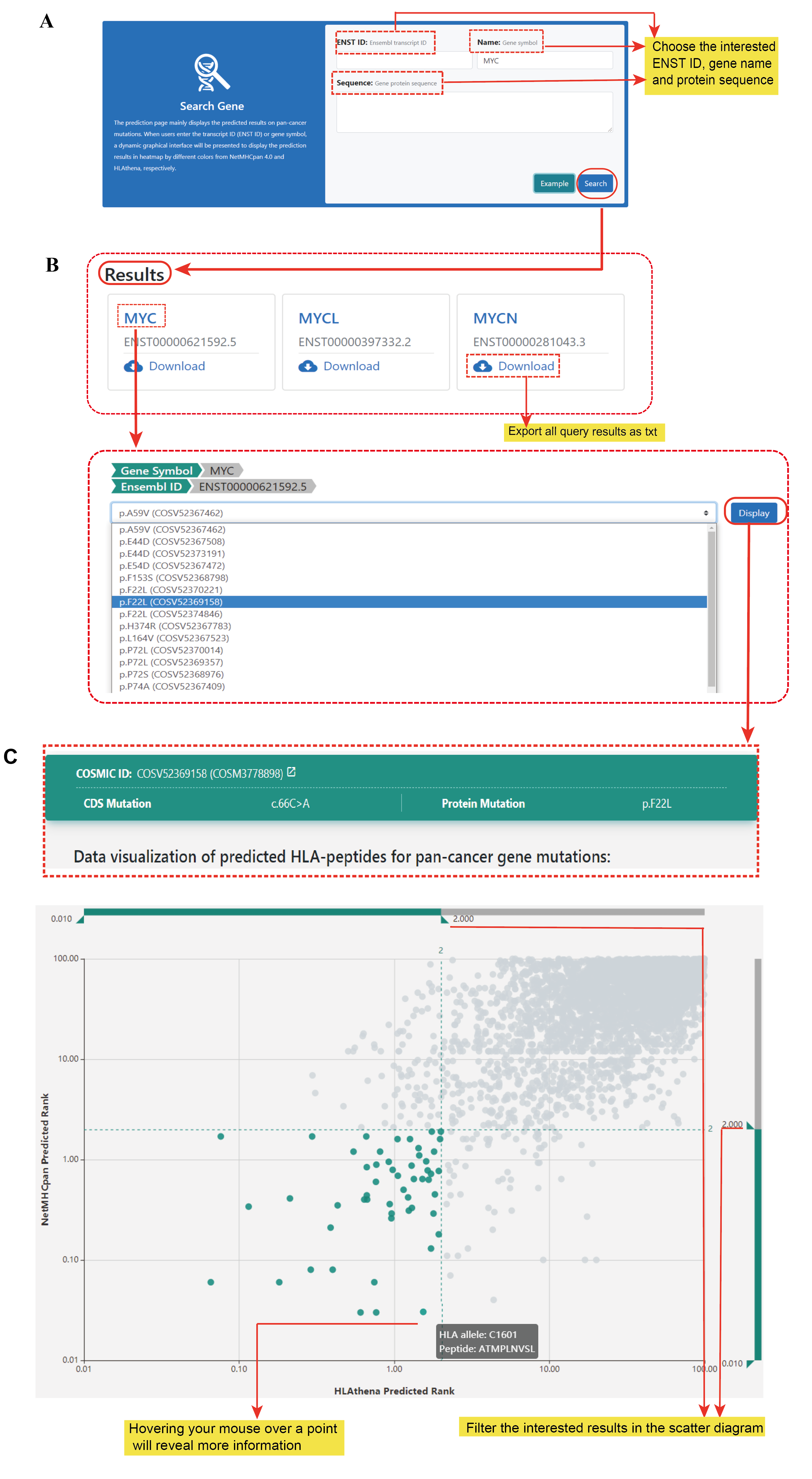
Each dot represents a binding between a peptide and a HLA I, plotted by predicted rank of HLAthena on the horizontal axis, and rank of netMHCpan on vertical. Both of them are base-10 logarithmic. You can filter the interested prediction results by pulling the barcodes on the upper and right sides. Points within the selected range are colored in blue. The closer the blue dot is to the lower left corner, the higher the probability that the peptide and HLA binding.
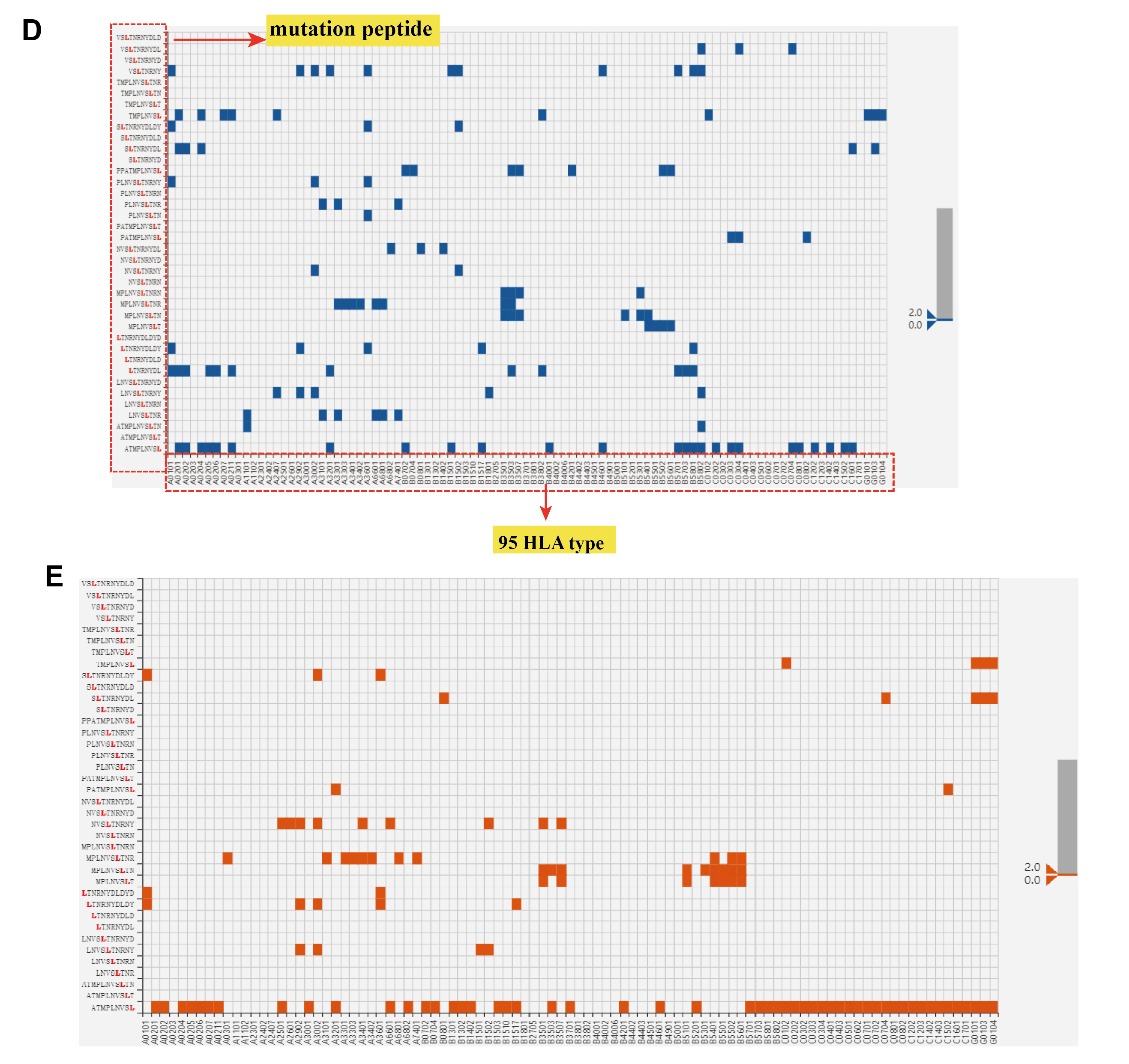
Data visualization of predicted HLA-peptides for pan-cancer gene mutations. (C) Dynamic scatter plot of predicted HLA-peptides (netMHCpan rank vs. HLAthena rank in log10 scale). The rank threshold can be adjusted to select reliable HLA-peptides highlighted in blue towards the lower left corner. (D) Heatmap showing the predicted bindings between peptides (left) and 95 HLA-A, -B, -C, and –G alleles (bottom) by NetMHCpan. The blue square indicates the HLA-peptides with rank less than 2. (E) Heatmap showing the predicted results by HLAthena.
How to use analysis tools?
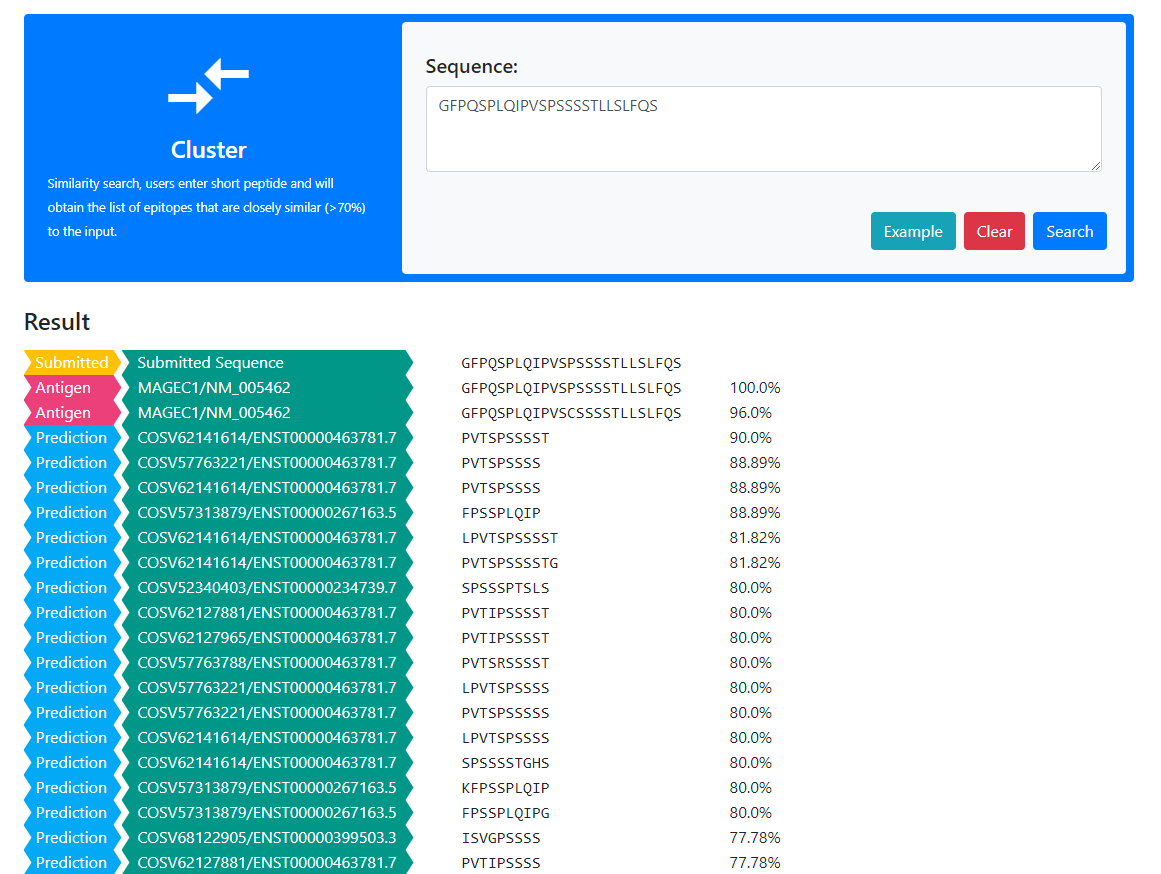
The web server mainly provides two tools: similarity search and subsequence search, which are used to search Validated Neopeptide Dataset (VND) and Predicted Neopeptide Dataset (PND)
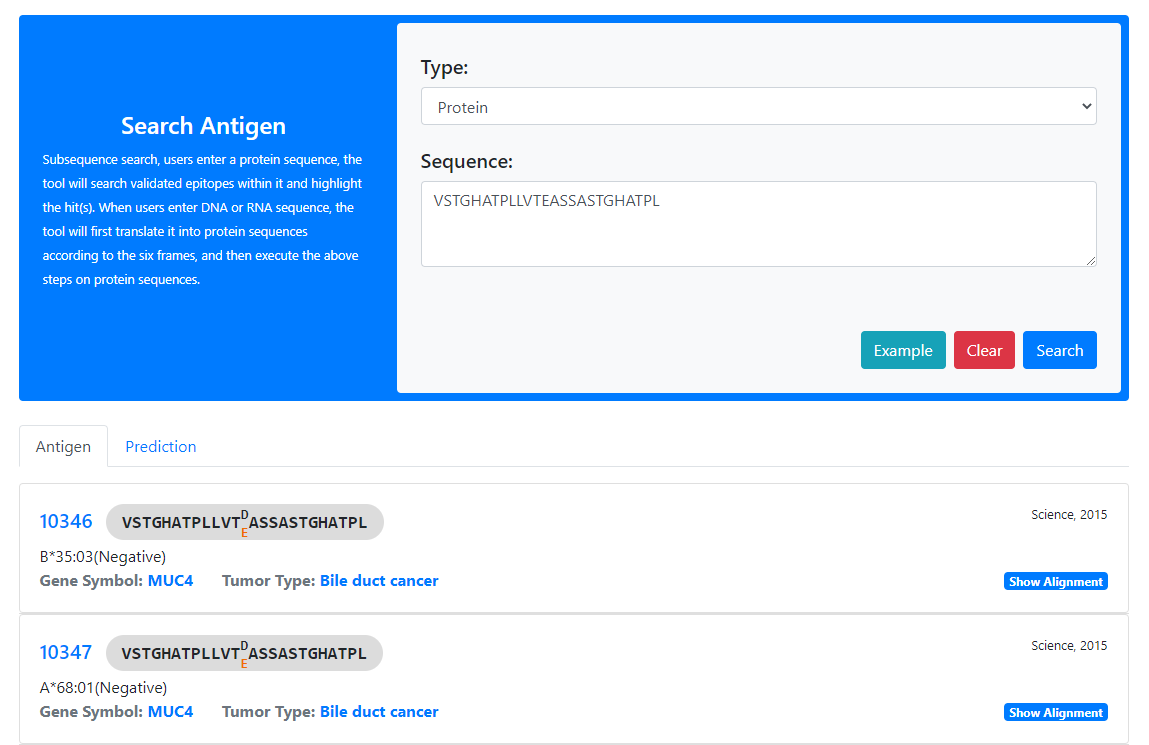
How to submit related data?
We support investigators to provide neoantigen data or to our database. Users can submit relevant neoantigens to our database in the following ways:
- Use the submit interface to submit new data.
- Send new data or publications to our email address: yu.zhou@whu.edu.cn or yinlei@whu.edu.cn. We will reply you as soon as possible.
Related resources
IEDB Immune Epitope Database Analysis Resource, http://www.iedb.org
NCBI National Center for Biotechnology Information, https://www.ncbi.nlm.nih.gov
MHCflurry 1.20, https://github.com/openvax/mhcflurry
NetMHC 4.0, http://www.cbs.dtu.dk/cgi-bin/sw_request?netMHC
NetMHCpan 4.0, http://www.cbs.dtu.dk/cgi-bin/sw_request?netMHCpan
NetMHCcons 1.1, http://www.cbs.dtu.dk/cgi-bin/sw_request?netMHCcons
HLAthena, http://hlathena.tools
PickPocket 1.1, http://www.cbs.dtu.dk/cgi-bin/sw_request?pickpocket
UCSC, https://genome.ucsc.edu
Ensembl, http://asia.ensembl.org/index.html
COSMIC, https://cancer.sanger.ac.uk/cosmic
National Cancer Institute, https://www.cancer.gov/types
GeneCards, https://www.genecards.org